Structure Creation and Representation
|
Palaios, G.A. and Hamodrakas, S.J.
|
 |
Index | |
 |
2.Copyright notice | |
 |
4. Description of the Output |
3. Input description
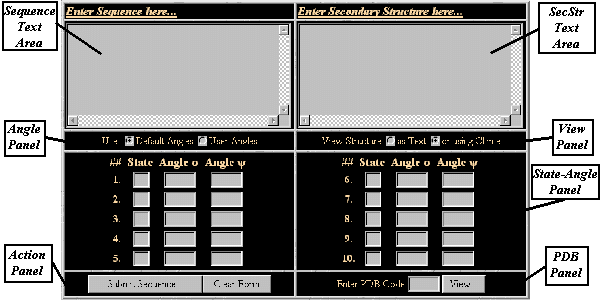
The input form, which allows a user to specify the data needed
by the program, is divided in seven panels:
- The Sequence of the polypeptide to be
created is specified in the Sequence text Area.
- The Secondary Structure is specified in the
SecStr text Area
- In the Angle Panel the user can either choose to
use a set of predefined phi,psi angles.
- In the View Panel the user can choose the
Output Mode by selecting either the
graphical output or the text output.
- If the 'User Angles' radio-button is checked, the user is forced to enter
in the State-Angles Panel the Letter of the Secondary Structure's
state and to define the phi, psi angles of the state.
- The 'Submit Sequence...' button in the Action Panel activates
the Create Module, whreas the 'Clear Form' button resets the input form.
- Finally in the PDB Panel the user can enter a PDB ID Code
and by pressing the 'View...' button activates the program that retrieves
the structure with the specified code, if exists, directly from the PDB Databank
- Errors appear in a new HTML Page.
A sequence can be either typed directly in the text area or
pasted from a text editor. The sequence can be given in FASTA
or Swiss-Prot Format. The Swiss-Prot Format contains
information that is not useful to the program so
it is recommended to use the FASTA format in order
to increase the reliability of the transmitted data and reduce the time of data transmission.
For convenience, it is also possible to enter a sequence as plain text
by using the one-letter code of aminoacid residues.
Spaces are allowed since they do not affect
the results of the program. The maximum length of a sequence is
2.000 residues(15.000 atoms).
The Secondary Structure can be either typed directly in the text area or
pasted from a text editor. If the user leaves the Secondary Structure Text
Area blanc, the program produces a polypeptide chain in an extended
form (phi, psi angles = 180). The SecStr
program of our Server can predict the secondary structure
of a polypeptide, SCAR can recognize only 4 letters of Secondary Structure
States(fig 1).
It is recommended to enter the Secondary Structure as plain text.
Spaces are allowed since they do not affect the results of the program.
SCAR's predefined angles are shown in the table below(Fig.1).
| ## | State Letter | phi angle | psi angle |
1 | H | -57 | -47 |
2 | E | -120 | +115 |
3 | B | -140 | +135 |
4 | L | -60 | -30 |
(fig 1)
- The 'H' letter is used to define a-helical structures
constist of a stretch of residues with phi = -57 psi = -47.
- The 'E' letter is used to define structures of parallel b-sheets
which consist of residues with phi = -120 and psi = +115.
- The 'B' letter is used to define structures of antiparallel b-sheets
which consist of residues with phi = -140 and psi = +135.
- The 'L' letter is used to define loops
which consist of residues with phi = -60 and psi = -30.
If the 'Default Angles' radio-button in the Angle Panel is checked, the
program uses the above predefined angles and state letters. If the 'User Angles'
is checked, then the user is obliged to input up to 10 states of secondary
structure with the ö and ø angles of each state specified at will.
Error messages are displayed in a new HTML Error Page.
Explanation of the error messages:
- Error 1 : No Sequence Entered.
- Error 2 : Check the Length of your Input.
- The length of the Sequence is different than the length of the Secondary Structure.
- Error 3 : Invalid Aminoacid Character.
- The Sequence entered contains letters that cannot be used. Allowed characters
are the 20 aminoacid letters and the Space character.
- Error 4 : Unrecognized Secondary Structure Character.
- The Secondary Structure either contains letters other than the 4 predefined('H', 'E', 'B', 'L') or
it contains characters other than those entered by the user.
- Error 5 : No Secondary Structure States entered.
3.5 Appendix
3.5.1 Input Form Layout
3.5.2 AminoAcids Letter Codes
| Aminoacid | Three Letter Code | One Letter Code |
| Alanine | Ala | A |
| Arginine | Arg | R |
| Asparagine | Asn | N |
| Aspartic Acid | Asp | D |
| Cysteine | Cys | C |
| Glutamine | Gln | Q |
| Glutamic Acid | Glu | E |
| Glycine | Gly | G |
| Histidine | His | H |
| Isoleucine | Ile | I |
| Leucine | Leu | L |
| Lysine | Lys | K |
| Methionine | Met | M |
| Phenylalanine | Phe | F |
| Proline | Pro | P |
| Serine | Ser | S |
| Threonine | Thr | T |
| Tryptophane | Trp | W |
| Tyrosine | Tyr | Y |
| Valine | Val | V |